I tried lots of ways, but i could not fix it. What you do with this. When there are ties, wilcox.test uses a normal approximation.
Chapter 6 From data visualization to statistical modelling A
You can see the code here:
Here is a slightly simplified version.
Stats.wilcoxon detects when d== 0, and appropriately warns that 'exact' will not work under such a circumstance. The default mode == 'auto' uses the exact distribution if there are at. My issue is about stats.wilcoxon(). (and default value is exact =.
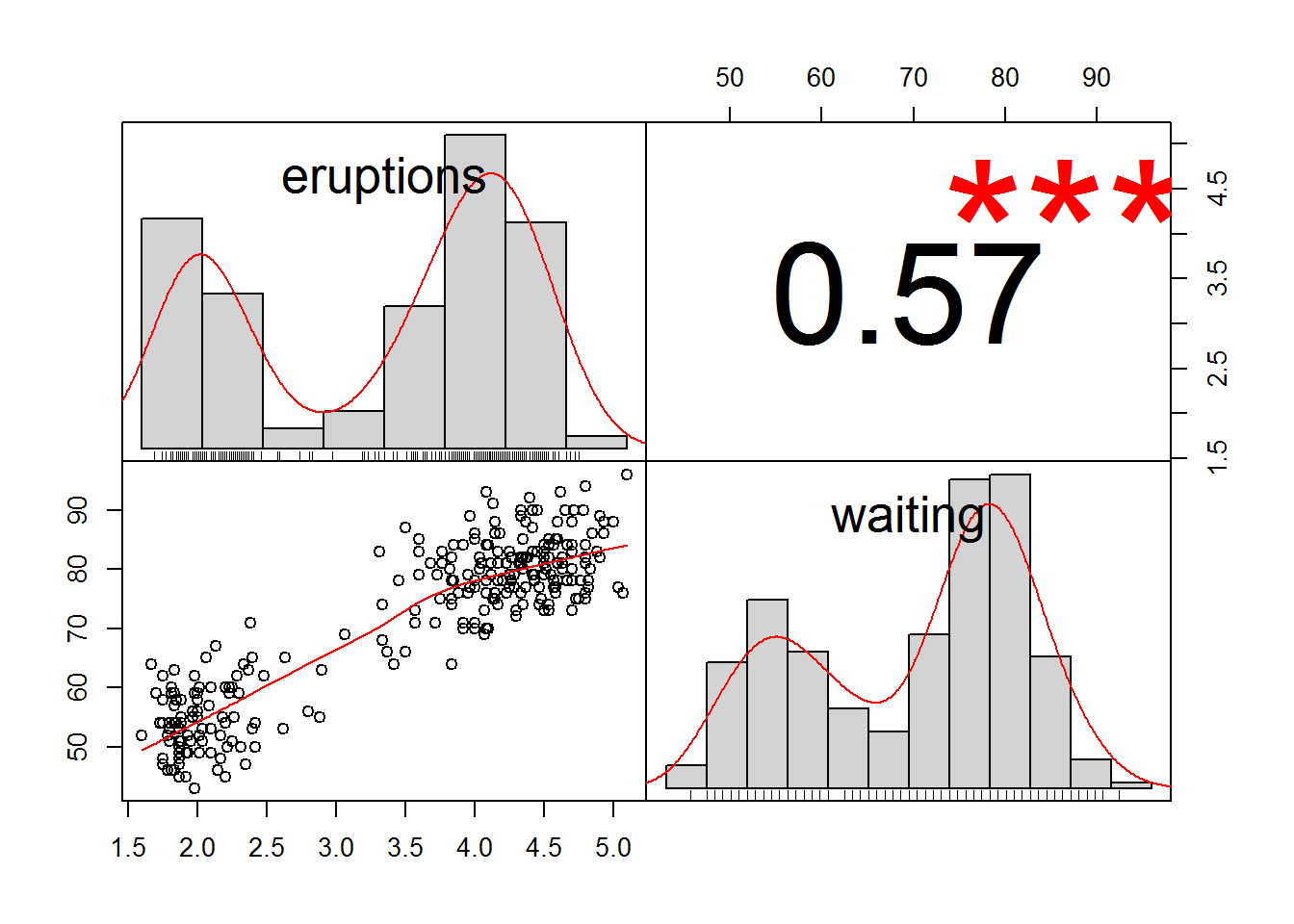
I'm running a spearman rank correlation on a data matrix and have the following code in rstudio: So this gives me the chart, as i asked for, but also gives me the warning. For nearly normal data, there is no difficulty using a t test.